library(tidyverse)
birth <- read_csv("datasets/birth.csv") # Remember to set the correct path
# Recoding sex as factor
birth <- birth |> mutate(sex = as.factor(ifelse(sex == 1, "Male", "Female")))
glimpse(birth)Rows: 230
Columns: 7
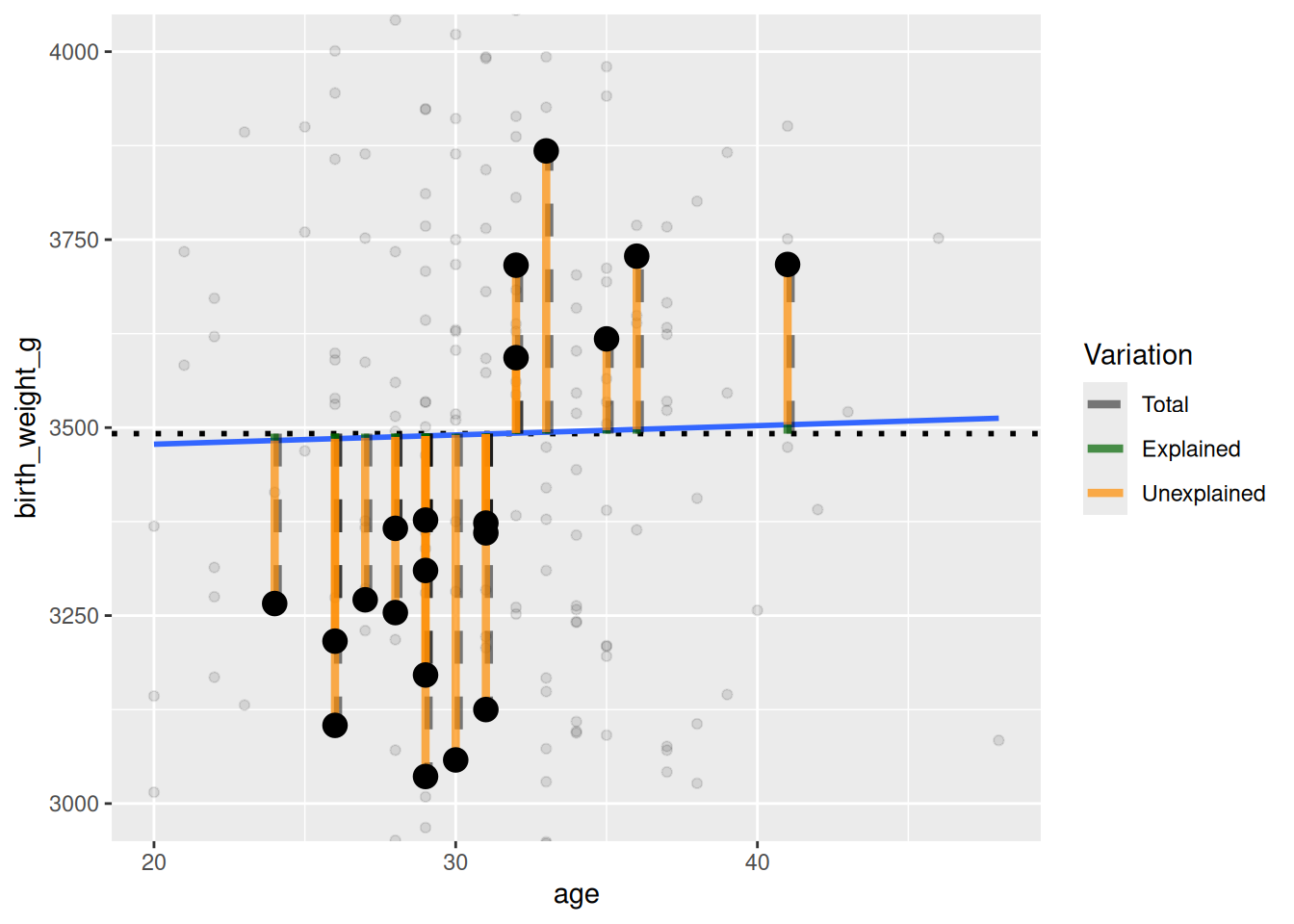
$ birth_weight_g <dbl> 4073, 4060, 3071, 3546, 3222, 3420, 3106, 3274,…
$ pregnancy_duration_wks <dbl> 41, 40, 39, 39, 39, 41, 40, 40, 40, 41, 41, 38,…
$ sex <fct> Male, Female, Female, Male, Female, Male, Femal…
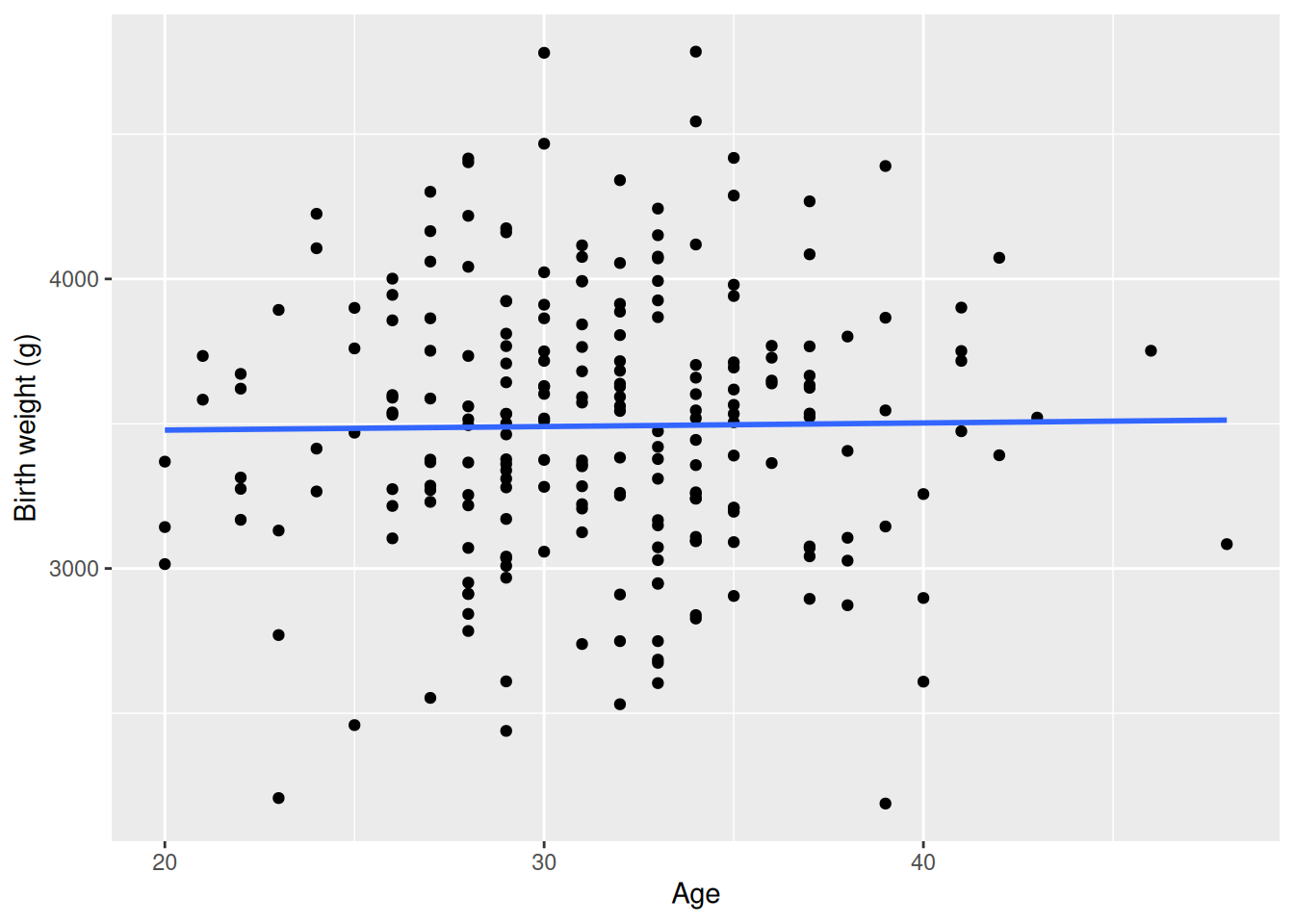
$ age <dbl> 42, 27, 37, 39, 31, 33, 38, 26, 30, 35, 29, 29,…
$ length_cm <dbl> 176, 180, 171, 167, 158, 166, 169, 164, 167, 16…
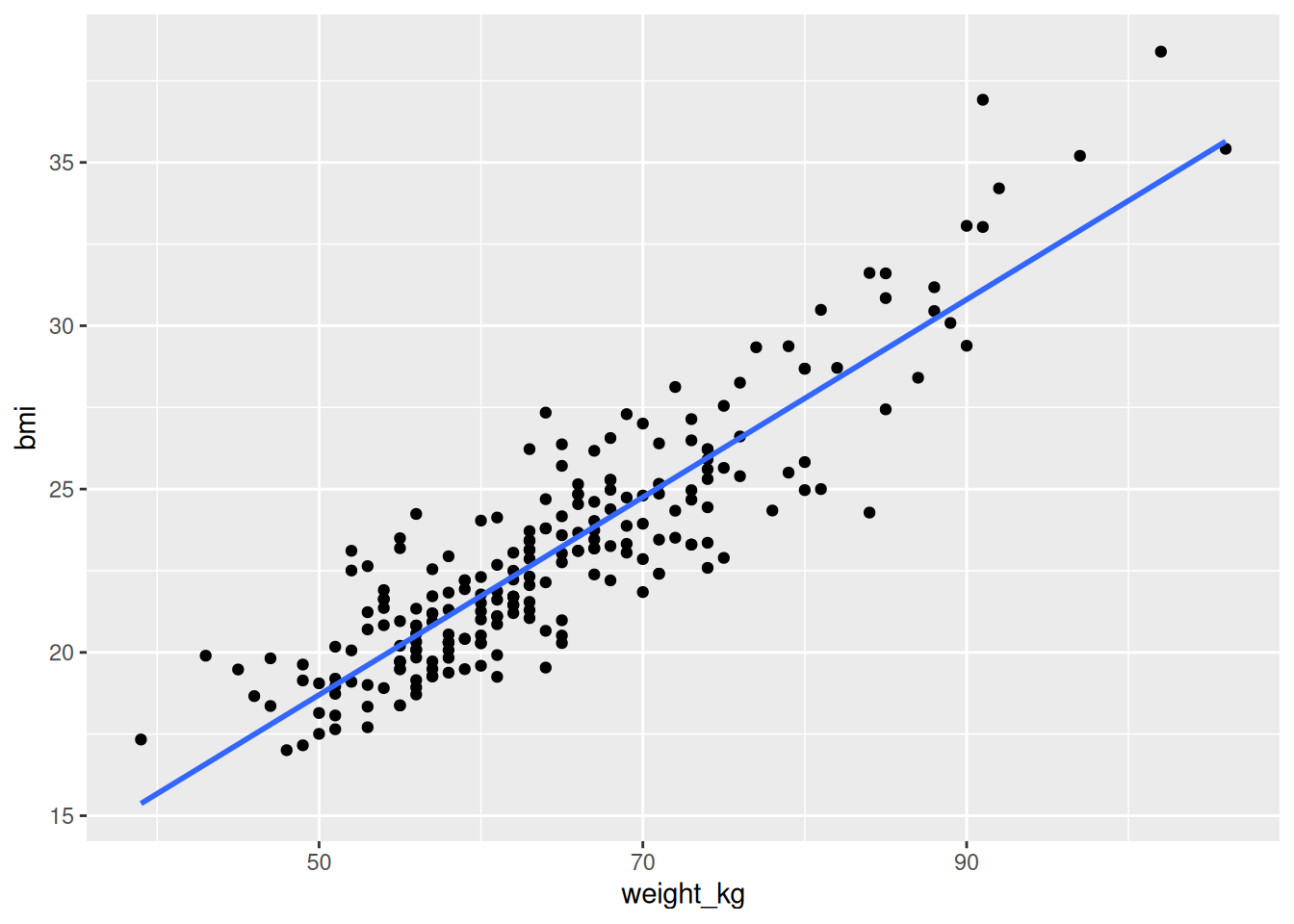
$ weight_kg <dbl> 65, 81, 70, 80, 54, 50, 50, 65, 80, 55, 57, 63,…
$ bmi <dbl> 20.98399, 25.00000, 23.93899, 28.68514, 21.6311…